Overview of eolearn.core
eolearn.core is the main subpackage which implements the basic building blocks:
EOPatch,EOTask,EONode,EOWorkflow,EOExecutor,
and commonly used functionalities.
EOPatch
The first basic object in the package is a data container, called EOPatch.
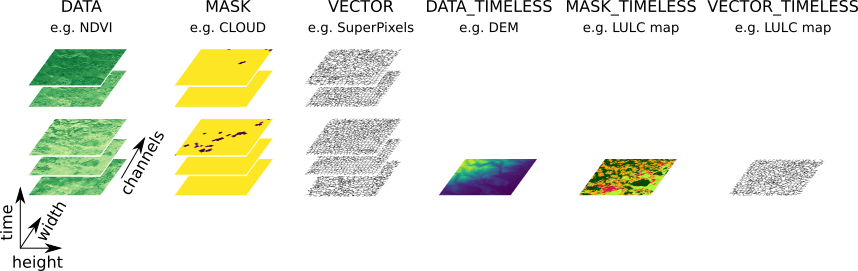
It is designed to store all types of EO data for a single geographical location.
The
EOPatchcan contain data (of the same location) for multiple times. If theEOPatchcontains multiple collections of temporal data, they must have the same temporal axis (the images must correspond to the same time-points).There is no limit to how much data a single
EOPatchcan store, but typically it shouldn’t be more than the size of your RAM.
Each EOPatch has an attribute bbox of type sentinelhub.BBox to define its area. The attribute timestamps defines the temporal component of an EOPatch, which is either None (for patches without a temporal dimension) or a list of datetime.datetime objects.
EO data can be divided into categories, called “feature types” according to the following properties:
|
Type of data |
Time component |
Spatial component |
Type of values |
Python object |
Shape |
|---|---|---|---|---|---|---|
DATA |
raster |
yes |
yes |
float |
|
|
MASK |
raster |
yes |
yes |
integer |
|
|
SCALAR |
raster |
yes |
no |
float |
|
|
LABEL |
raster |
yes |
no |
integer |
|
|
DATA_TIMELESS |
raster |
no |
yes |
float |
|
|
MASK_TIMELESS |
raster |
no |
yes |
integer |
|
|
SCALAR_TIMELESS |
raster |
no |
no |
float |
|
|
LABEL_TIMELESS |
raster |
no |
no |
integer |
|
|
VECTOR |
vector |
yes |
yes |
/ |
|
Required columns |
VECTOR_TIMELESS |
vector |
no |
yes |
/ |
|
Required column |
META_INFO |
anything |
no |
no |
anything |
anything |
anything |
Note: t specifies time component, n and m are spatial components (height and width), and d is an additional component for data with multiple channels.
Let’s start by loading an existing EOPatch and displaying it’s content (i.e. features):
[1]:
import os
from eolearn.core import EOPatch
INPUT_FOLDER = os.path.join("..", "..", "example_data")
INPUT_EOPATCH = os.path.join(INPUT_FOLDER, "TestEOPatch")
eopatch = EOPatch.load(
INPUT_EOPATCH, lazy_loading=False # Set this parameter to True to load data in memory only when first needed
)
eopatch
[1]:
EOPatch(
bbox=BBox(((465181.0522318204, 5079244.8912012065), (466180.53145382757, 5080254.63349641)), crs=CRS('32633'))
timestamps=[datetime.datetime(2015, 7, 11, 10, 0, 8), ..., datetime.datetime(2017, 12, 22, 10, 4, 15)], length=68
mask_timeless={
LULC: numpy.ndarray(shape=(101, 100, 1), dtype=uint16)
RANDOM_UINT8: numpy.ndarray(shape=(101, 100, 13), dtype=uint8)
VALID_COUNT: numpy.ndarray(shape=(101, 100, 1), dtype=int64)
}
vector={
CLM_VECTOR: geopandas.GeoDataFrame(columns=['TIMESTAMP', 'VALUE', 'geometry'], length=55, crs=EPSG:32633)
}
label={
IS_CLOUDLESS: numpy.ndarray(shape=(68, 1), dtype=bool)
RANDOM_DIGIT: numpy.ndarray(shape=(68, 2), dtype=int8)
}
meta_info={
maxcc: 0.8
service_type: 'wcs'
size_x: '10m'
size_y: '10m'
}
scalar_timeless={
LULC_PERCENTAGE: numpy.ndarray(shape=(6,), dtype=float64)
}
scalar={
CLOUD_COVERAGE: numpy.ndarray(shape=(68, 1), dtype=float16)
}
vector_timeless={
LULC: geopandas.GeoDataFrame(columns=['index', 'RABA_ID', 'AREA', 'DATE', 'LULC_ID', 'LULC_NAME', 'geometry'], length=88, crs=EPSG:32633)
}
mask={
CLM: numpy.ndarray(shape=(68, 101, 100, 1), dtype=uint8)
CLM_INTERSSIM: numpy.ndarray(shape=(68, 101, 100, 1), dtype=bool)
CLM_MULTI: numpy.ndarray(shape=(68, 101, 100, 1), dtype=bool)
CLM_S2C: numpy.ndarray(shape=(68, 101, 100, 1), dtype=bool)
IS_DATA: numpy.ndarray(shape=(68, 101, 100, 1), dtype=uint8)
IS_VALID: numpy.ndarray(shape=(68, 101, 100, 1), dtype=bool)
}
label_timeless={
LULC_COUNTS: numpy.ndarray(shape=(6,), dtype=int32)
}
data_timeless={
DEM: numpy.ndarray(shape=(101, 100, 1), dtype=float32)
MAX_NDVI: numpy.ndarray(shape=(101, 100, 1), dtype=float64)
}
data={
BANDS-S2-L1C: numpy.ndarray(shape=(68, 101, 100, 13), dtype=float32)
CLP: numpy.ndarray(shape=(68, 101, 100, 1), dtype=float32)
CLP_MULTI: numpy.ndarray(shape=(68, 101, 100, 1), dtype=float32)
CLP_S2C: numpy.ndarray(shape=(68, 101, 100, 1), dtype=float32)
NDVI: numpy.ndarray(shape=(68, 101, 100, 1), dtype=float32)
}
)
There are multiple ways how to access a feature in the EOPatch.
[2]:
from eolearn.core import FeatureType
# All of these access the same feature:
bands = eopatch.data["BANDS-S2-L1C"]
# or
bands = eopatch[FeatureType.DATA]["BANDS-S2-L1C"]
# or
bands = eopatch[(FeatureType.DATA, "BANDS-S2-L1C")]
# or
bands = eopatch[FeatureType.DATA, "BANDS-S2-L1C"]
type(bands), bands.shape
[2]:
(numpy.ndarray, (68, 101, 100, 13))
Vector features are handled by geopandas:
[3]:
eopatch[FeatureType.VECTOR, "CLM_VECTOR"].head()
[3]:
| TIMESTAMP | VALUE | geometry | |
|---|---|---|---|
| 0 | 2015-07-31 10:00:09 | 1.0 | POLYGON ((465181.052 5080254.633, 465181.052 5... |
| 1 | 2015-08-20 10:07:28 | 1.0 | POLYGON ((465181.052 5080254.633, 465181.052 5... |
| 2 | 2015-09-19 10:05:43 | 1.0 | POLYGON ((465181.052 5080254.633, 465181.052 5... |
| 3 | 2015-09-29 10:06:33 | 1.0 | POLYGON ((465181.052 5080254.633, 465181.052 5... |
| 4 | 2015-12-08 10:04:09 | 1.0 | POLYGON ((465181.052 5080254.633, 465181.052 5... |
Special features are bounding box and timestamps:
[4]:
print(eopatch.timestamps[:5])
print(repr(eopatch.bbox))
eopatch.bbox.geometry # draws the shape of BBox
[datetime.datetime(2015, 7, 11, 10, 0, 8), datetime.datetime(2015, 7, 31, 10, 0, 9), datetime.datetime(2015, 8, 20, 10, 7, 28), datetime.datetime(2015, 8, 30, 10, 5, 47), datetime.datetime(2015, 9, 9, 10, 0, 17)]
BBox(((465181.0522318204, 5079244.8912012065), (466180.53145382757, 5080254.63349641)), crs=CRS('32633'))
[4]:
A list of all features in an EOPatch can be obtained with:
[5]:
eopatch.get_features()
[5]:
[(<FeatureType.DATA: 'data'>, 'CLP_S2C'),
(<FeatureType.DATA: 'data'>, 'CLP'),
(<FeatureType.DATA: 'data'>, 'NDVI'),
(<FeatureType.DATA: 'data'>, 'BANDS-S2-L1C'),
(<FeatureType.DATA: 'data'>, 'CLP_MULTI'),
(<FeatureType.MASK: 'mask'>, 'CLM'),
(<FeatureType.MASK: 'mask'>, 'IS_DATA'),
(<FeatureType.MASK: 'mask'>, 'CLM_MULTI'),
(<FeatureType.MASK: 'mask'>, 'CLM_INTERSSIM'),
(<FeatureType.MASK: 'mask'>, 'IS_VALID'),
(<FeatureType.MASK: 'mask'>, 'CLM_S2C'),
(<FeatureType.SCALAR: 'scalar'>, 'CLOUD_COVERAGE'),
(<FeatureType.LABEL: 'label'>, 'IS_CLOUDLESS'),
(<FeatureType.LABEL: 'label'>, 'RANDOM_DIGIT'),
(<FeatureType.VECTOR: 'vector'>, 'CLM_VECTOR'),
(<FeatureType.DATA_TIMELESS: 'data_timeless'>, 'DEM'),
(<FeatureType.DATA_TIMELESS: 'data_timeless'>, 'MAX_NDVI'),
(<FeatureType.MASK_TIMELESS: 'mask_timeless'>, 'RANDOM_UINT8'),
(<FeatureType.MASK_TIMELESS: 'mask_timeless'>, 'LULC'),
(<FeatureType.MASK_TIMELESS: 'mask_timeless'>, 'VALID_COUNT'),
(<FeatureType.SCALAR_TIMELESS: 'scalar_timeless'>, 'LULC_PERCENTAGE'),
(<FeatureType.LABEL_TIMELESS: 'label_timeless'>, 'LULC_COUNTS'),
(<FeatureType.VECTOR_TIMELESS: 'vector_timeless'>, 'LULC'),
(<FeatureType.META_INFO: 'meta_info'>, 'maxcc'),
(<FeatureType.META_INFO: 'meta_info'>, 'size_x'),
(<FeatureType.META_INFO: 'meta_info'>, 'size_y'),
(<FeatureType.META_INFO: 'meta_info'>, 'service_type')]
Let’s create a new EOPatch and store some features inside.
[6]:
import numpy as np
from sentinelhub import CRS, BBox
# Since EOPatch represents geolocated data, it should always have a bounding box
new_eopatch = EOPatch(bbox=BBox((0, 0, 1, 1), CRS.WGS84))
new_eopatch[FeatureType.MASK_TIMELESS, "NEW_MASK"] = np.zeros((68, 10, 13), dtype=np.uint8)
# If temporal features are added to an EOPatch that does not have timestamps (or if the dimensions do not match),
# the user is warned that the EOPatch is temporall ill-defined
new_eopatch.timestamps = eopatch.timestamps
new_eopatch[FeatureType.DATA, "BANDS"] = eopatch[FeatureType.DATA, "BANDS-S2-L1C"]
# The following wouldn't work as there are restrictions to what kind of data can be stored in each feature type
# new_eopatch[FeatureType.MASK, 'NEW_MASK'] = np.zeros((10, 10, 13), dtype=np.uint8)
# new_eopatch[FeatureType.VECTOR, 'NEW_MASK'] = np.zeros((10, 10, 13), dtype=np.uint8)
new_eopatch
[6]:
EOPatch(
bbox=BBox(((0.0, 0.0), (1.0, 1.0)), crs=CRS('4326'))
timestamps=[datetime.datetime(2015, 7, 11, 10, 0, 8), ..., datetime.datetime(2017, 12, 22, 10, 4, 15)], length=68
mask_timeless={
NEW_MASK: numpy.ndarray(shape=(68, 10, 13), dtype=uint8)
}
data={
BANDS: numpy.ndarray(shape=(68, 101, 100, 13), dtype=float32)
}
)
It is also possible to delete a feature:
[7]:
del new_eopatch[FeatureType.MASK_TIMELESS, "NEW_MASK"]
new_eopatch
[7]:
EOPatch(
bbox=BBox(((0.0, 0.0), (1.0, 1.0)), crs=CRS('4326'))
timestamps=[datetime.datetime(2015, 7, 11, 10, 0, 8), ..., datetime.datetime(2017, 12, 22, 10, 4, 15)], length=68
data={
BANDS: numpy.ndarray(shape=(68, 101, 100, 13), dtype=float32)
}
)
We can save EOPatch into a local folder. In case an EOPatch already exists in the specified location, we have to allow to overwrite its features.
[8]:
from eolearn.core import OverwritePermission
OUTPUT_FOLDER = os.path.join(".", "outputs")
os.makedirs(OUTPUT_FOLDER, exist_ok=True)
NEW_EOPATCH_PATH = os.path.join(OUTPUT_FOLDER, "NewEOPatch")
new_eopatch.save(NEW_EOPATCH_PATH, overwrite_permission=OverwritePermission.OVERWRITE_FEATURES)
Let’s load the saved version and compare it with original:
[9]:
loaded_eopatch = EOPatch.load(NEW_EOPATCH_PATH)
new_eopatch == loaded_eopatch
[9]:
True
Each EOPatch can be shallow or deep copied:
[10]:
new_eopatch.copy()
new_eopatch.copy(deep=True)
[10]:
EOPatch(
bbox=BBox(((0.0, 0.0), (1.0, 1.0)), crs=CRS('4326'))
timestamps=[datetime.datetime(2015, 7, 11, 10, 0, 8), ..., datetime.datetime(2017, 12, 22, 10, 4, 15)], length=68
data={
BANDS: numpy.ndarray(shape=(68, 101, 100, 13), dtype=float32)
}
)
EOTask
The next core object is EOTask, which is a single well-defined operation on one or more EOPatch objects.
We can create a new EOTask by creating a class that inherits from the abstract EOTask class:
class FooTask(EOTask):
def __init__(self, foo_param):
""" Task-specific parameters
"""
self.foo_param = foo_param
def execute(self, eopatch, *, patch_specific_param):
# Do what foo does on EOPatch and return it
return eopatch
In the initialization method we define task-specific parameters.
Each task has to implement the
executemethod.executemethod has to be defined in a way that:positional arguments have to be instances of
EOPatch,other types of arguments should be keyword arguments.
Otherwise the task itself can do anything.
Example of a task that adds a new feature to existing EOPatch:
[11]:
from typing import Any, Tuple
from eolearn.core import EOTask
class AddFeatureTask(EOTask):
"""Adds a feature to the given EOPatch.
:param feature: Feature to be added
:type feature: (FeatureType, feature_name) or FeatureType
"""
def __init__(self, feature: Tuple[FeatureType, str]):
self.feature = feature
def execute(self, eopatch: EOPatch, *, data: Any) -> EOPatch:
"""Returns the EOPatch with added features.
:param eopatch: input EOPatch
:param data: data to be added to the feature
:return: input EOPatch with the specified feature
"""
eopatch[self.feature] = data
return eopatch
Let’s see how such a task could be used.
[12]:
eopatch = EOPatch(bbox=BBox((0, 0, 1, 1), CRS.WGS84), timestamps=[f"2017-0{i}-01" for i in range(1, 6)])
add_feature_task = AddFeatureTask((FeatureType.DATA, "NEW_BANDS"))
data = np.zeros((5, 100, 100, 13))
eopatch = add_feature_task.execute(eopatch, data=data)
eopatch
[12]:
EOPatch(
bbox=BBox(((0.0, 0.0), (1.0, 1.0)), crs=CRS('4326'))
timestamps=[datetime.datetime(2017, 1, 1, 0, 0), ..., datetime.datetime(2017, 5, 1, 0, 0)], length=5
data={
NEW_BANDS: numpy.ndarray(shape=(5, 100, 100, 13), dtype=float64)
}
)
The majority of eo-learn consists of different EOTasks implementing different operations on EO data.
The list of all EOTasks is available in the documentation.
EONode and EOWorkflow
EOTasks can be joined together into an acyclic processing graph called EOWorkflow. Since eo-learn 1.0 these tasks first have to be wrapped into instances of EONode class.
Here is a simple example of how an EOWorkflow can be created:
[13]:
from eolearn.core import EONode, EOWorkflow, LoadTask, SaveTask
new_feature = FeatureType.LABEL, "NEW_LABEL"
load_task = LoadTask(path=INPUT_FOLDER)
add_feature_task = AddFeatureTask(new_feature)
save_task = SaveTask(path=OUTPUT_FOLDER, overwrite_permission=OverwritePermission.OVERWRITE_FEATURES)
# Each EONode object defines dependecies to other EONode objects:
load_node = EONode(load_task, inputs=[], name="Load EOPatch")
add_feature_node = EONode(add_feature_task, inputs=[load_node], name="Add a new feature")
save_node = EONode(save_task, inputs=[add_feature_node], name="Save EOPatch")
workflow = EOWorkflow([load_node, add_feature_node, save_node])
# or
workflow = EOWorkflow.from_endnodes(save_node)
# Alternatively, a linear workflow could also be built with a helper function:
# from eolearn.core import linearly_connect_tasks
# nodes = linearly_connect_tasks(load_task, add_feature_task, save_task)
# workflow = EOWorkflow(nodes)
Let’s display the dependency graph:
[14]:
%matplotlib inline
workflow.dependency_graph()
[14]:
EOWorkflow is executed by specifying EOPatch-related parameters:
[15]:
results = workflow.execute({
load_node: {"eopatch_folder": "TestEOPatch"},
add_feature_node: {"data": np.zeros((68, 3), dtype=np.uint8)},
save_node: {"eopatch_folder": "WorkflowEOPatch"},
})
results
[15]:
WorkflowResults(outputs={}, start_time=datetime.datetime(2023, 8, 28, 15, 19, 54, 733751), end_time=datetime.datetime(2023, 8, 28, 15, 19, 54, 961589), stats={'LoadTask-939b27aa45a511eeb8db-91a8de8b81da': NodeStats(node_uid='LoadTask-939b27aa45a511eeb8db-91a8de8b81da', node_name='Load EOPatch', start_time=datetime.datetime(2023, 8, 28, 15, 19, 54, 733806), end_time=datetime.datetime(2023, 8, 28, 15, 19, 54, 822464), exception_info=None), 'AddFeatureTask-939b2a9b45a511eea69d-e2612971e907': NodeStats(node_uid='AddFeatureTask-939b2a9b45a511eea69d-e2612971e907', node_name='Add a new feature', start_time=datetime.datetime(2023, 8, 28, 15, 19, 54, 825206), end_time=datetime.datetime(2023, 8, 28, 15, 19, 54, 825267), exception_info=None), 'SaveTask-939b2cb545a511eea722-ed1665ca815d': NodeStats(node_uid='SaveTask-939b2cb545a511eea722-ed1665ca815d', node_name='Save EOPatch', start_time=datetime.datetime(2023, 8, 28, 15, 19, 54, 827230), end_time=datetime.datetime(2023, 8, 28, 15, 19, 54, 960678), exception_info=None)}, error_node_uid=None)
A result of a workflow execution is a WorkflowResults object. It contains information about times of each node execution and information about potential errors.
Note:
A difference between executing tasks directly and executing tasks in a workflow is that in a workflow each EOPatch input object will be first shallow-copied before being passed to any task.
EOExecutor
EOExecutor handles the execution and monitoring of EOWorkflows. It enables executing a workflow multiple times and in parallel. At the end, it generates a report containing the summary of the workflow’s execution process.
Execute previously defined workflow with different arguments.
[16]:
from eolearn.core import EOExecutor
execution_args = [ # EOWorkflow will be executed for each of these 5 dictionaries:
{
load_node: {"eopatch_folder": "TutorialEOPatch"},
add_feature_node: {"data": idx * np.ones((10, 3), dtype=np.uint8)},
save_node: {"eopatch_folder": f"ResultEOPatch{idx}"},
}
for idx in range(5)
]
executor = EOExecutor(workflow, execution_args, save_logs=True, logs_folder=OUTPUT_FOLDER)
results = executor.run(workers=3) # The execution will use at most 3 parallel processes
100%|██████████| 5/5 [00:00<00:00, 510.50it/s]
Make the report:
[17]:
executor.make_report()
print(f"Report was saved to location: {executor.get_report_path()}")
Report was saved to location: /home/ubuntu/Sinergise/eo-learn/examples/core/outputs/eoexecution-report-2022_02_09-12_38_30/report.html